NEWS: Pcons is now supported by the EGI FedCloud with the VO VO.NBIS.SE for computational resource.
".
NEWS: Pcons now use the sum of S-score instead of average LGscore.
This means that the Pcons score essentially reflects the "number of correct residues".
Pcons is a Model Quality Assessment Program (MQAP) which means that it ranks protein models by assessing their quality. Pcons uses a set of possible protein models as input. These models can, and should, be produced using various methods and approaches. The Pcons protocol analyzes the set of protein models and looks for recurring three-dimensional structural patterns. Based on this analysis each model is assigned a score reflecting how common its three-dimensional structural patterns are in the whole ensemble of models. The idea being that recurring patterns are more likely to be correct as compared to patterns than only occur in one or just a few models.
The result from the Pcons protocol is one score reflecting the overall global quality, and a score for each individual residue in the protein reflecting the local residue quality.
Pcons was the best Model Quality Assessment Program in CASP7, Assilomar 2007 and CASP8, Cagliari 2008. Please observe the curves below refer to the old scoring scheme using LGscore.
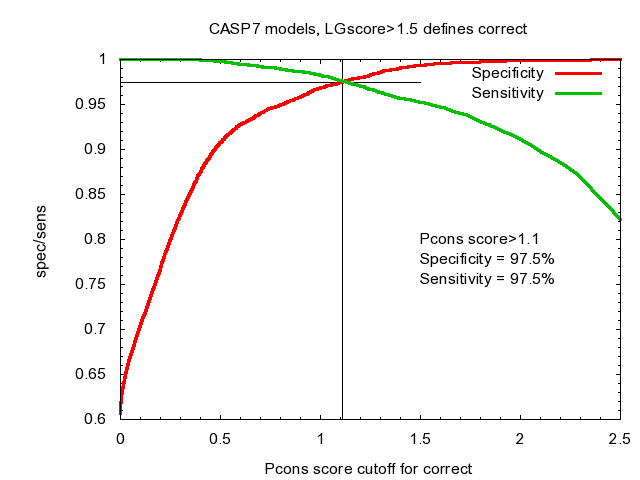
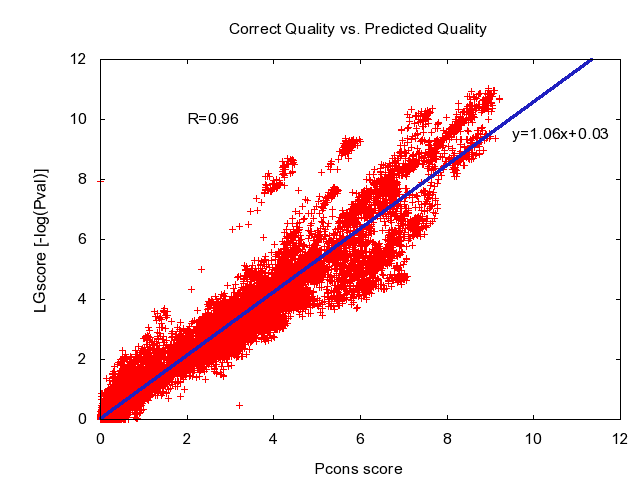
Automatic consensus based fold recognition using Pcons, ProQ and Pmodeller. Björn Wallner, Huisheng Fang and Arne Elofsson (2003). Proteins. 53 Suppl 6:534-541.
Pcons5: combining consensus, structural evaluation and fold recognition scores. Björn Wallner and Arne Elofsson (2005). Bioinformatics., 21(23):4248-4254.
Identification of correct regions in protein models using structural, alignment and consensus information. Björn Wallner and Arne Elofsson (2006). Protein Sci., 15(4):900-913.
|
|
|
|